ClinCode - Computer-Assisted Clinical ICD-10 Coding for improving efficiency and quality in healthcare
Hercules Dalianis a, Taridzo Chomutare b, Andrius Budrionis b, Therese Olsen Svenning b
Introduction
Manual ICD-10 diagnosis coding is time consuming and error prone (up to 20-30 percent errors) according to studies in Sweden and Norway. Lots of unstructured data is stored digitally in large Electronic Patient Records systems. Simultaneously, Natural Language Processing and Artificial Intelligence methods have shown their efficiency in various text processing tasks, such as text classification, topic modeling, machine translation, text summarisation and others. These factors combined have made it possible to create a Computer-Assisted Clinical Coding (CAC) tool for ICD-10 codes that suggest codes to the discharge summary.
However, a number of challenges still exist, especially for minor languages like Swedish and Norwegian, primarily driven by inaccessibility of clinical data for training data-driven algorithms. Deep learning methods using large language models are investigated as potentially useful tools for training CAC models. The objective of the current study was to document the feasibility of predicting ICD-10 blocks from Swedish discharge summaries, towards a future pipeline designed to predict the full code.
Methods
The data contains 6,062 Swedish discharge summaries from 4,985 unique patients with 263 unique ICD-10 codes codes in the Gastro surgery medical speciality. The data is divided into ten logical blocks: K00-K14, K20-K31 up to K90-K93. The data have been used to fine tune a clinical language deep learning BERT model called SweClin-BERT encompassing two million patients.
To obtain better training data for ICD-10 coding a set of Norwegian 4,799 discharge summaries have been re-coded with the correct ICD-10 codes. This data set will also be used to predict ICD-10 codes.
Results
The results show an F1-score of 0.835 for the prediction of ICD-10 at 10 block level using the clinical language model SweClin-BERT (Lamproudis et al. 2022).
A web based demo called ICD-10 coder1 has been created to demonstrate the code assignment. see Figure 1. However that model is based on KB-BERT a general Swedish Language model that does not contain sensitive personal information, (Remmer et al. 2021).
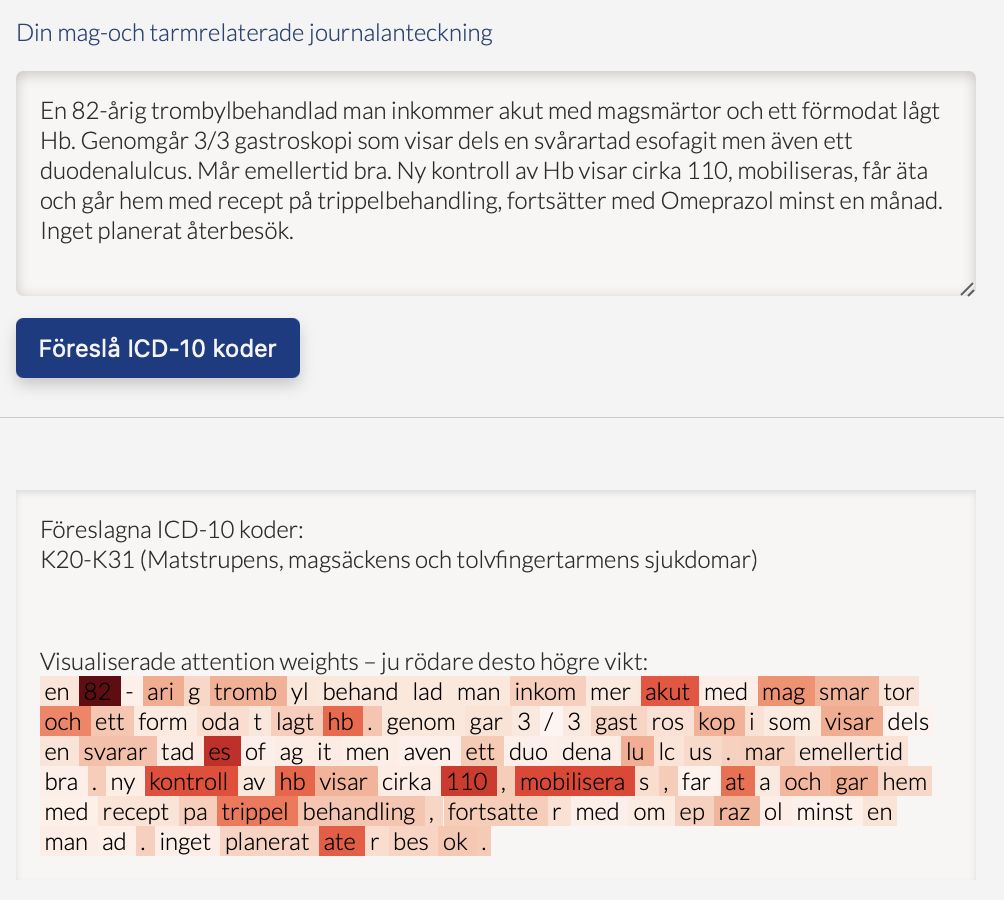
Fig 1. The web based ICD-10 coder.
Current coding practice is error-prone. An analysis of all discharge summaries from a Norwegian Gastro Clinic over the course of 3 years (3,554 patients) showed an Inter Annotator Agreement in Kappa value of 0.66 for the whole code-line and 0.7 for the main diagnosis isolated. Kappa values are not directly translated to percentages, but the results correspond to a moderate agreement where 35-63% of the data are reliable. A large portion of errors came from misregistration of complications.
Conclusions
It is shown that automatic assignment of ICD-10 codes is feasible using machine learning and creates plausible results at least at 10 block level.
Prediction at the block-level was an important preliminary step in the project. Experiments are continuing, using recent advances in deep learning and fuzzy logic, to improve results and specifically in predicting unique full codes, as well as developing the mechanism for explaining which words in the discharge summary give the highest weight for the prediction of a specific ICD-10 code.
References
- https://icdcoder.dsv.su.se
- Remmer, S., Lamproudis, A. and H Dalianis. 2021. Multi-label Diagnosis Classification of Swedish Discharge Summaries - ICD-10 Code Assignment Using KB-BERT. In the Proceedings of RANLP 2021: Recent Advances in Natural Language Processing, 1-3 Sept 2021, Varna, Bulgaria,
- Lamproudis, A., Henriksson, A. and H. Dalianis. 2022. Vocabulary Modifications for Domain-adaptive Pretraining of Clinical Language Models. In Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies - Volume 5: HEALTHINF, pp. 180-188
a DSV-Stockholm University, Sweden
b National Centre for E-health Research, Norway
Original Version in PDF
